Notes For All Chapters Biology Class 12 CBSE
The DNA and RNA World:
1. Over the years after Mendel, the nature of the genetic material was investigated, resulting in the realisation that DNA is the genetic material in majority of organisms.
2. Deoxyribonucleic acid (DNA) and Ribonucleic acid (RNA) are the two types of nucleic acid found in living systems. Nucleic acids are polymers of nucleotides.
3. DNA acts as a genetic material in most organisms, whereas RNA acts as a genetic material in some viruses.
4. RNA mostly functions as messenger. RNA has other functions as adapter, structural or as a catalytic molecule.
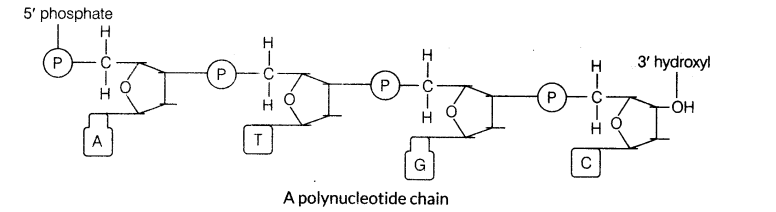
5. Structure of Polynucleotide Chain
(i) A nucleotide has three parts, i.e. a nitrogenous base, a pentose sugar (deoxyribose in DNA and ribose in RNA) and a phosphate group.
(ii) Nitrogenous bases are purines, i.e. adenine, guanine and pyrimidines, i.e. cytosine, uracil and thymine.
(iii) Cytosine is common for both DNA and RNA and thymine is present in DNA. Uracil is present in RNA at the place of thymine.
(iv) A nitrogenous base is linked to the pentose sugar through a N-glycosidic linkage to form a nucleoside, i.e. adenosine and guanosine, etc.
(v) When a phosphate group is linked to 5′ —OH of a nucleoside through phosphodiester linkage, a corresponding nucleotide is formed.
(vi) Two nucleotides are linked through 3′ -> 5′ phosphodiester linkage to form a dinucleotide.
(vii) Several nucleotides can be joined to form a polynucleotide chain.
(viii) The backbone in a polynucleotide chain is formed due to sugar and phosphates.
(ix) The nitrogenous bases linked to sugar moiety project from the backbone.
(x) The base pairs are complementary to each other.
6. In case of RNA, every nucleotide residue has an additional—OH group present at 2-position in the ribose. Also, the uracil is found at the place of thymine (5-methyl uracil).
7. Discoveries Related to Structure of DNA
(i) Friedrich Meischer in 1869, first identified DNA as an acidic substance present in the nucleus and named it as ‘nuclein’.
(ii) James Watson and Francis Crick, proposed a very simple double helix model for the structure of DNA in 1953 based on X-ray diffraction data.
(iii) Erwin Chargaff proposed that for a double-stranded DNA, the ratios between adenine and thymine and guanine and cytosine are constant and equals to one.
8. Salient Features of Double-helix Structure of DNA
(i) DNA is a long polymer of deoxyribonucleotides. It is made up of two polynucleotide chains, where the backbone is constituted by sugar-phosphate and the bases project inside.
(ii) The two chains have anti-parallel polarity, i.e. 5′ > 3′ for one, 3′ > 5′ for another.
(iii) The bases in two strands are paired through hydrogen bond (H—bonds) forming base pairs (bp). Adenine forms two hydrogen bonds with thymine from opposite strand and vice-versa. Guanine bonds with cytosine by three H—bonds. Due to this, purine always comes opposite to a pyrimidine. This forms a uniform distance between the two strands.
(iv) The two chains are coiled in a right-handed fashion. The pitch of the helix is 3.4 nm and there are roughly 10 bp in each turn. Due to this, the distance between a base pair in a helix is about 0.34 nm.
(v) The plane of one base pair stacks over the other in double helix. This confers stability to the helical structure in addition to H—bonds.
9. The length of a DNA double helix is about 2.2 meters (6.6 x 109 bp x 0.34 x 10-9 m/bp)
Therefore, it needs special packaging in a cell.
(i) In prokaryotic cells (which do not have a defined nucleus), such as E.coli, DNA (being negatively charged) is held with some proteins (that have positive charges) in a region called as nucleoid. The DNA in nucleoid is organised in large loops held by proteins.
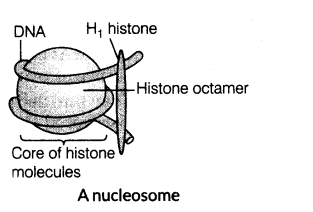
(ii) In eukaryotes, there is a set of positively charged proteins called histones that are rich in basic amino acid residues, lysines and arginines (both positive).
Histones are organised to form a unit of eight molecules called histone octamer. The negatively charged DNA is wrapped around the positively charged histone octamer to form a structure called nucleosome.
(iii) A typical nucleosome contains 200 bp of DNA helix.
Nucleosomes constitute the repeating unit of a structure in nucleus called chromatin (thread-like stained structure). Under electron microscope, the nucleosomes in chromatin can be seen as beads-on-string. This structure in chromatin is packaged to form chromatin fibres that further coils and condense to form chromosomes at metaphase stage.
(iv) The packaging of chromatin at higher level requires additional set of proteins which are collectively called Non-Histone Chromosomal (NHC) proteins.
(v) In a nucleus, some regions of chromatin are loosely packed (stains light) called euchromatin (transcriptionally active chromatin). In some regions, chromatin is densely packed (stains dark) called heterochromatin (inactive chromatin).
10. Transfonning Principle
(i) Frederick Griffith (1928) carried out a series of experiments with Streptococcus pneumoniae (bacterium causing pneumonia).
(ii) According to him, when the bacteria are grown on a culture plate, some produce smooth shiny colonies (S), while others produce rough (R) colonies.
(iii) This is because the S-strain bacteria have a mucous (polysaccharide) coat, while R-strain does not.
(iv) Mice infected with S-strain (virulent) die from pneumonia but mice infected with R-strain do not develop pneumonia.
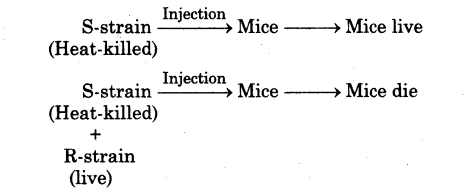
(v) Griffith killed bacteria by heating and observed that heat-killed S-strain bacteria injected into mice did not kill them. On injecting mixture of heat-killed S and live R bacteria, the mice died. He recovered living S-bacteria from dead mice.
(vi) From this experiment, he concluded that the ‘R-strain bacteria’ had been transformed by the heat-killed S-strain bacteria. Some transforming principle transferred from heat-killed S-strain, had enabled the R-strain to synthesise a smooth polysaccharide coat and become virulent. This must be due to transfer of the genetic material.
11. Biochemical Nature of Transforming Principle
(i) Oswald Avery, Colin MacLeod and Maclyn McCarty, worked to determine the biochemical nature of transforming principle in Griffith’s experiment.
(ii) They purified biochemicals (proteins, RNA and DNA, etc) from heat-killed S-cells and discovered that DNA alone from S-bacteria caused R-bacteria to be transformed.
(iii) They also discovered that protease (protein digesting enzyme) and RNAases (RNA-digesting enzymes) did not affect transformation.
(iv) Digestion with DNAse did inhibit transformation, indicating that DNA caused transformation.
(v) They concluded that DNA is the hereditary material. But, still all the biologists were not convinced.
12. DNA is the Genetic Material
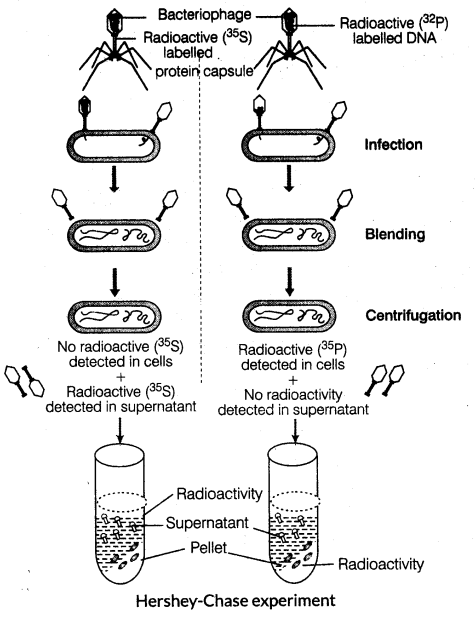
(i) Alfred Hershey and Martha Chase (1952) gave unequivocal proof that DNA is the genetic material.
(ii) In their experiments, bacteriophages (viruses that infect bacteria) were used.
(iii) They grew some viruses on a medium that contained radioactive phosphorus and some others on sulphur containing radioactive medium.
(iv) Viruses grown in the presence of radioactive phosphorus contained radioactive DNA but not radioactive protein because DNA contains phosphorus but protein does not. In the same way, viruses grown on radioactive sulphur contained radioactive protein, but not radioactive DNA because DNA does not contain sulphur.
(v) Radioactive phages were allowed to attach to E. coli bacteria. As the infection proceeded, viral coats were removed from the bacteria by agitating them in a blender. The virus particles were separated from the bacteria by spinning them in a centrifuge.
(vi) Bacteria which were infected with viruses that had radioactive DNA were radioactive, indicating that DNA was the material that passed from the virus to the bacteria.
(vii) Bacteria that were infected with viruses that had radioactive proteins were not radioactive. This indicated that the proteins did not enter the bacteria from viruses. It proved that DNA is a genetic material that is passed from virus to bacteria.
13. Properties of Genetic Material
(i) It became establised that DNA is the genetic material from the Hershey-Chase experiment.
(ii) In some viruses, RNA was also reported as genetic material, e.g. Tobacco mosaic viruses, QB bacteriophage, etc.
(iii) Characteristics of a Genetic Material
(a) It should be able to replicate.
(b) It should be chemically and structurally stable.
(c) It should provide scope for slow changes (mutation) that are required for evolution.
(d) It should be able to express itself in the form of ‘Mendelian characters’.
(iv) According to the above mentioned rules, both the nucleic acids (DNA and RNA) have the ability to direct duplications.
Stability can be explained in DNA as the two strands being complementary if separated by heating come together in appropriate conditions.
(v) The 2′ — OH group present at every nucleotide in RNA is a reactive group and makes RNA labile and easily degradable, hence it is reactive.
(vi) DNA is chemically less reactive and structurally more stable when compared to RNA. Thymine also confers additional stability to DNA. So, among the two nucleic acids, the DNA is a predominant genetic material.
(vii) Both RNA and DNA are able to mutate. Viruses having RNA genome and having shorter life span mutate and evolve faster.
(viii) DNA is dependent on RNA for protein synthesis, while RNA can directly code for it. The protein synthesising machinery has evolved around RNA. This concluded that the DNA being more stable is suitable for storage of genetic information, while for the transmission of genetic information, RNA is suitable.
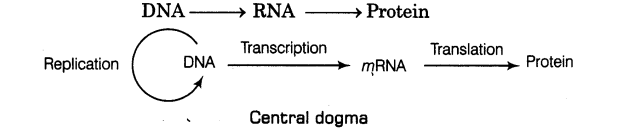
14. Francis Crick proposed the central dogma in molecular biology, which states that the
genetic information flows from
15. Replication Scheme for replication of DNA termed as semiconservative DNA replication was proposed by Watson and Crick (1953). According to it,
(i) The two strands would separate and act as a template for the synthesis of new
complementary strands. .
(ii) After replication, each DNA molecule would have one parental and one newly synthesised strand.
16. Experimental proof that DNA replicates semiconservatively, comes first from E. coli and later from higher organisms, such as plants and human cells.
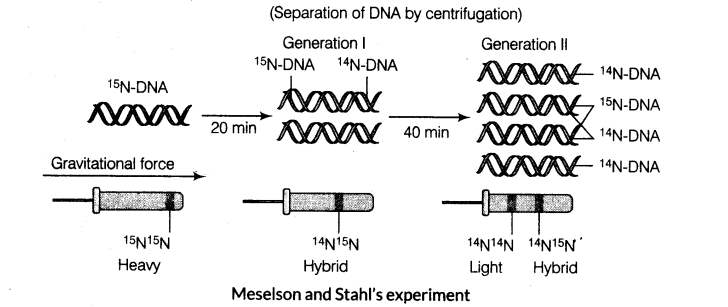
Matthew Meselson and Franklin Stahl performed the following experiments to prove this in 1958.
(i) E. coli was grown in a medium containing 15NH4C1 as the only nitrogen source for many generations. 15N got incorporated into newly synthesised DNA (and other nitrogen containing compounds). This heavy DNA molecule could be distinguished from the normal DNA by centrifugation in a cesium chloride (CsCl) density gradient.
(ii) They then transferred the cells into a medium with normal 14NH4Cl and took samples at various definite intervals as the cells multiplied and extracted the DNA that remained as double stranded helices. DNA samples were separated independently on CsCl gradients to measure DNA densities.
(iii) The DNA that was extracted from the culture, one generation (after 20 min) after the transfer from 15 N to 14N medium had a hybrid or intermediate density. DNA extracted from the culture after another generation (after 40 min) was composed of equal amounts of this hybrid DNA and of light DNA.
(iv) Very similar experiments were carried out by Taylor and Colleagues on Vicia faba (faba beans) using radioactive thymidine and the same results, i.e. DNA replicates semiconservatively, were obtained as in earlier experiments.
17. DNA replication machinery and enzymes process of replication requires a set of catalysts (enzymes).
(i) The main enzyme is DNA-dependent DNA polymerase, since it uses a DNA template to catalyse the polymerisation of deoxynucleotides. The average rate of polymerisation by these enzymes is approximately 2000 bp/second.
(ii) These polymerases has to catalyse the reaction with high degree of accuracy because any mistake during replication would result into mutations.
DNA polymerisation is an energy demanding process, so deoxyribonucleoside triphosphates serve dual purposes, i.e. act as substrates and provide energy for polymerisation reaction.
(iv) Many additional enzymes are also required in addition to DNA-dependent DNA polymerase.
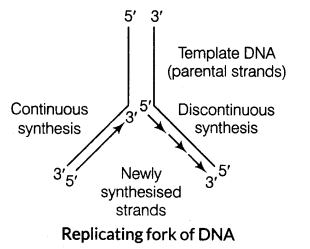
(v) (a) Replication in DNA strand occurs within a small opening of the DNA helix, known as replication fork.
(b) DNA-dependent DNA polymerases catalyse polymerisation only in one direction, i.e. 5′ -> 3. It creates additional complications at the replicating fork. Consequently, on one strand (template 3′-5′), the replication is continuous, while on the other strand (template 5′-3′), it is discontinuous. The discontinuously synthesised fragments called Okazaki fragments are later joined by DNA ligase.
18. Origin of Replication
(i) DNA polymerases cannot initiate the process of replication on their own. Also, replication does not initiate randomly at any place in DNA. So, there is a definite region in E.coli DNA where the replication originates. The region is termed as origin of replication.
(ii) Due to this requirement, a piece of DNA, if needed to be propagated during recombinant DNA procedures, requires a vector. The vectors provide the origin of replication.
19. RNA world RNA was the first genetic material. There are evidences to prove that essential life processes, such as metabolism, translation, splicing, etc., have evolved around RNA.
(i) There are some important biochemical reactions in living systems that are catalysed by RNA catalysts and not by protein enzyne.
(ii) DNA has evolved from RNA with chemical modifications that make it more stable because RNA being a catalyst was reactive and hence, unstable.
20. There are following three types of RNAs:
(i) mRNA (messenger RNA) provides the template for transcription.
(ii) tRNA (transfer RNA) brings amino acids and reads the genetic code.
(iii) rRNA (ribosomal RNA) plays structural and catalytic role during translation.
All the three RNAs are needed to synthesise a protein in a cell.
21. Traription is the process of copying genetic information from one strand of the i DNJS. into RNA. The principle of complementarity governs the process of
transcription, except the adenosine now forms base pair with uracil instead of l thymine.
(i) In transcription, only a segment of DNA is duplicated and on Iv one of the strands is . copied into RNA. Both the strands are not copied because
• If both the strands code for RNA, two different RNA me’ ^cules and two different proteins would be formed, hence complicating the genetic information transfer machinery.
• Since two RNA produced would be complementary to each other, they would form a double-stranded RNA without translation, making the process of transcription futile.
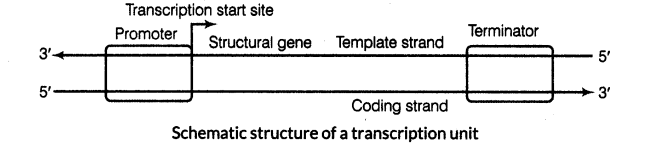
(ii) A transcription unit in DNA is defined by three regions in the DNA which are as follows:
(a) A promoter (b) The structural gene (c) A terminator
(iii) The two strands of DNA have opposite polarity and the DNA-dependent RNA polymerase also catalyse the polymerisation in only one direction that is 5′ -» 3′.
(iv) The strand that has the polarity 3′-» 5′ acts as a template and is referred to as template strand. The other strand which has the polarity (5′ -> 3′) and the sequence same as RNA (T at the place of U) is displaced during transcription. This strand is called as coding strand.
(v) The promoter and terminator flank the structural gene in a transcription unit.
(vi) The promoter is located towards 5′ end (upstream) of the structural gene.
(vii) It is the DNA sequence that provides binding site for RNA polymerase and the presence of promoter defines the template and coding strands. By switching its position with terminator, the definition of coding and template strands could be reversed.
(viii) The terminator is located towards 3f-end (downstream) of the coding strand and it usually defines the end of the process of transcription.
(ix) There are additional regulatory sequences that may be present further upstream or downstream to the promoter.
Transcription Unit and the Gene
(i) A gene can be defined as the functional unit of inheritance.
(ii) A cistron is a segment of DNA coding for a polypeptide.
(iii) The structural gene in a transcription unit could be said as monodstronic (mostly in eukaryotes) or polycistronic (mostly in bacteria or prokaryotes).
(iv) The coding sequences or expressed sequences are defined as exons. Exons appear in mature or processed RNA. The exons are interrupted by introns.
(v) Introns or intervening sequences do not appear in mature or processed RNA.
(vi) Sometimes, the regulatory sequences are loosely defined as regulate 5: even
though these sequences do not code for any RNA or protein.
22. Transcription in prokaryotes occur in the following steps:
(i) A single DNA-dependent RNA polymerase catalyses the transcription of all types of RNA in bacteria.
(ii) RNA polymerase binds to promotor and initiates transcription (initiation).
(iii) It uses nucleoside triphosphates as substrate and polymerises in a template depended fashion following the rule of complementarity.
It also facilitates opening of the helix and continues elongation.
(v) Once the polymerase reaches the terminator region, the nascent RNA falls off, so also the RNA polymerase. This results in termination of transcription.
(vi) RNA polymerase is only capable of catalysing the process of elongation.
It associates transiently with initiation-factor (a) and terminator factor (b), to initiate and terminate the transcription, respectively. Thus, catalysing all the three steps.
(vii) Since, the mRNA does not require any processing to become active and also since transcription and translation take place in the same compartment, many times the translation can begin much before the mRNA is fully transcribed. As a result, transcription and translation can be coupled in bacteria.
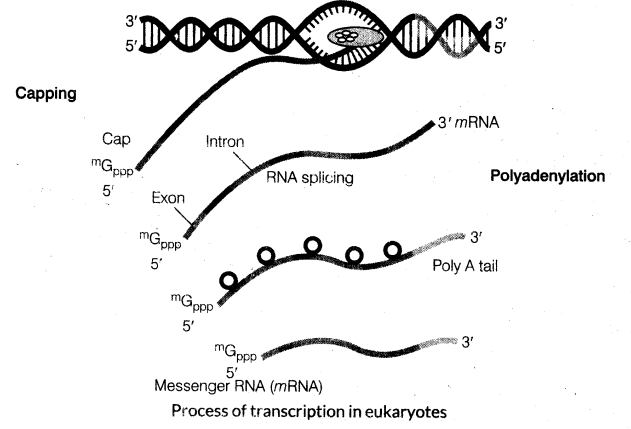
23. Transcription in eukaryotes have additional complexities than prokaryotes.
(i) There are at least three RNA polymerases in the nucleus other than the RNA polymerase in organelles. The RNA polymerase I transcribes rRNAs (28S, 18S and 5.8S). RNA polymerase III is responsible for transcription of fRNA, 5srRNA and SnRNAs (small nuclear RNAs). RNA polymerase II transcribes precursor of mRNA, the heterogenous nuclear RNA (/mRNA).
(ii) Another complexity is that, the primary transcripts contain both the exons and the introns and are non-functional. Hence, subject to a process called splicing. In this process, introns are removed and exons are joined in a definite order.
(iii) /mRNA undergoes additional processing called as capping and tailing. In capping, an unusual nucleotide is added to the 5′-end of /mRNA. In tailing, adenylate residues
(200-300) are added at 3′-end in a template. It is the fully processed /mRNA, now called mRNA, that is transported out of the nucleus for translation process.
Significance of these complexities are:
(i) The split gene arrangements represent an ancient feature of genome.
(ii) The presence of introns is reminescent of antiquity.
(iii) The process of splicing represents the dominance of RNA world.
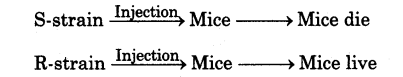
Leave a Reply